Analysis for Disease Gene Association using ML
Objective
The main objective of this application is to investigate a specific problem of whether it is valuable or not to use machine learning techniques to predict the type of Disease gene.
Abstract
To recognize the basis of disease, it is essential to determine its underlying genes. Understanding the association between underlying genes and genetic disease is a fundamental problem regarding human health. Identification and association of genes with the disease require time consuming and expensive experimentations of a great number of potential candidate genes. Therefore, the alternative inexpensive and rapid computational methods have been proposed that can identify the candidate gene associated with a disease. Most of these methods use phenotypic similarities due to the fact that genes causing same or similar diseases have less variation in their sequence or network properties of protein-protein interactions based on-premises that genes lie closer in protein interaction network that causes the similar or same disease. However, these methods use only basic network properties or topological features and gene sequence information or biological features as a prior knowledge for identification of gene-disease association, which restricts the identification process to a single gene-disease association. In this study, we propose and analyze some novel computational methods for the identification of genes associated with diseases. Some advance topological and biological features that are overlooked currently are introducing for identifying candidate genes. We evaluate different computational methods on disease-gene association data from DisGeNET based on TP rate, FP rate, precision, recall, F-measure, and ROC curve evaluation parameters. The results reveal that various computational methods with advanced feature set outperform previous state-of-the-art techniques by achieving precision up to 93.8%, recall up to 93.1%, and F- measure up to 92.9%. Significantly, we apply our methods to study three major disease types: Group, Disease and Phenotype. Simulation results sho44w that the proposed Extreme Gradient Boosting Algorithm (XGBoost) gives more accurate results as compared to previously published approaches.
KEYWORDS: Machine Learning, XGBoost Classifier, Disease Gene Association
NOTE: Without the concern of our team, please don't submit to the college. This Abstract varies based on student requirements.
Block Diagram
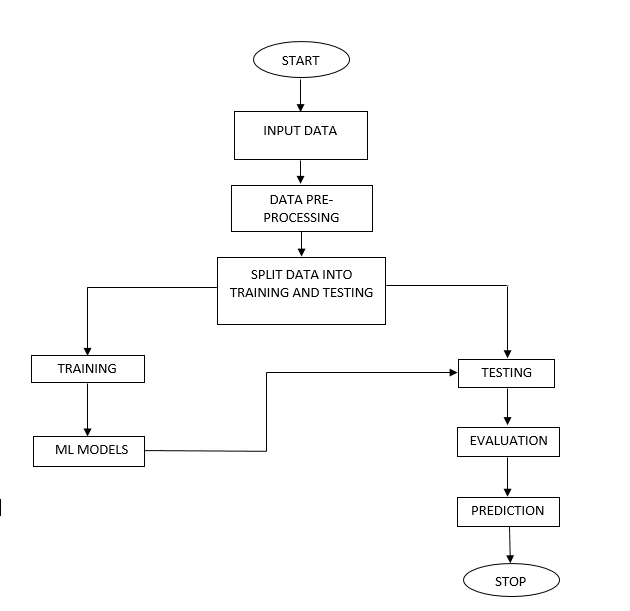
Specifications
HARDWARE SPECIFICATIONS:
- Processor- I3/Intel Processor
- RAM- 4GB (min)
- Hard Disk- 128 GB
- Key Board-Standard Window
- Keyboard. Mouse-Two or Three Button Mouse.
- Monitor-Any.
SOFTWARE SPECIFICATIONS:
- Operating System: Windows 7+
- Technology: Python 3.6+
- IDE: PyCharm IDE
- Libraries Used: Pandas, NumPy, Scikit-Learn, Matplotlib.
Learning Outcomes
- About Python.
- About Jupyter Notebook.
- About Pandas.
- About Numpy.
- About HTML.
- About CSS.
- About JavaScript.
- About Database.
- About Machine Learning.
- About Artificial Intelligent.
- About how to use the libraries.
- Cloud Overview.
- Terminology of cloud.
- Project Development Skills:
- Problem analyzing skills.
- Problem solving skills.
- Creativity and imaginary skills.
- Programming skills.
- Deployment.
- Testing skills.
- Debugging skills.
- Project presentation skills.
- Thesis writing skills.





 Paper Publishing
Paper Publishing
